Tutorial
1. General information
ProMENDA is a comprehensive database for depression. We hope that you benefit from this database for depression research. Please contact us with any comments or questions.
1) What is ProMENDA?
• ProMENDA is a comprehensive database that integrated all available knowledge of metabolomics, magnetic resonance spectroscopy and proteomics studies for depression.
2) What types of information does ProMENDA contain?
• Metabolomic database: All available differential metabolites integrated from more than 1,000 metabolomics and magnetic resonance spectroscopy studies in the field of depression.
• Proteomic database: All available differential proteins integrated from more than 200 proteomic studies in the field of depression.
• Web browser: A web browser containing browse and search function is provided, which could provide biological information from systematic level.
3) How the information was collected and organized in ProMENDA?
All data were manually collected and integrated by experienced researchers. The detail process of data collection, data extraction and data annotation are provided in the Introduction.
2. Browse
In the Browse module, both molecule-level and study-level information are provided.
1) The molecule-level information
In this part, metabolite or protein entries are listed alphabetically. For each entry, the information of molecule name, study ID, study type, organism, category of depression, tissue, platform, and up-/down-regulated direction are provided. The following functions are provided in the browse page (Figure 1).
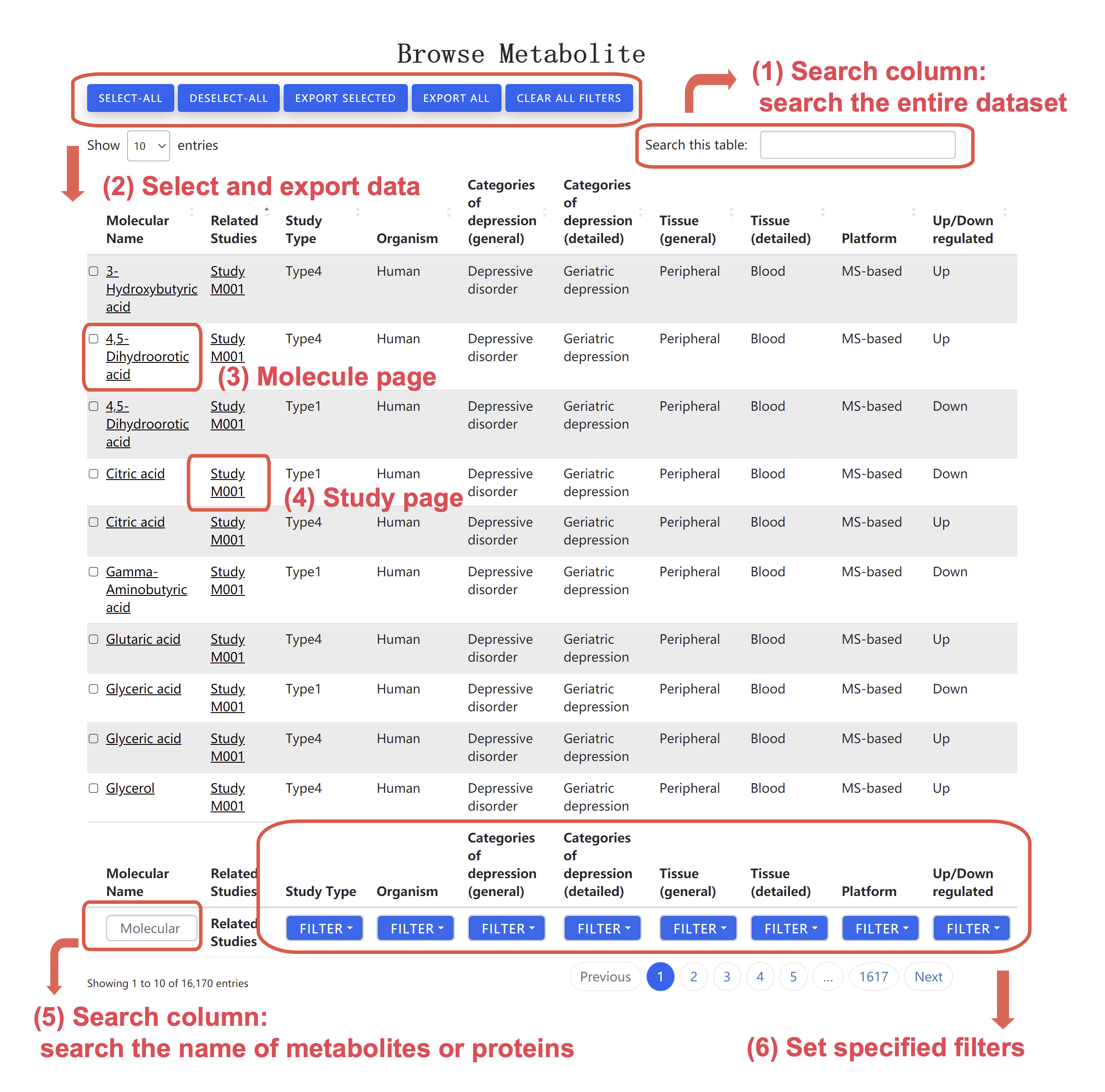
Figure 1. Snapshot of the search page.
• Search: Users can (i) search the entire dataset using the search column provide at the at the upper right corner, or (ii) search metabolite names (for metabolite data) or gene symbols (for protein data) using the search column provide at the at the lower left corner.
• Select and download data: Users can use the blue button in the upper left corner to select data of interest. In addition, users can download the selected data in Text format, and then open the document using Excel.
• Filter: Users can filter data of interest based on study type, organism, category of depression, tissue, platform, and up-/down-regulated direction.
• Molecule page: Users can click the name of the molecule, and then jump to the specified page for this molecule. In the specified page (Figure 2), external links and synonyms for the molecule are provided. Moreover, molecule entries are summarized and listed in the page.
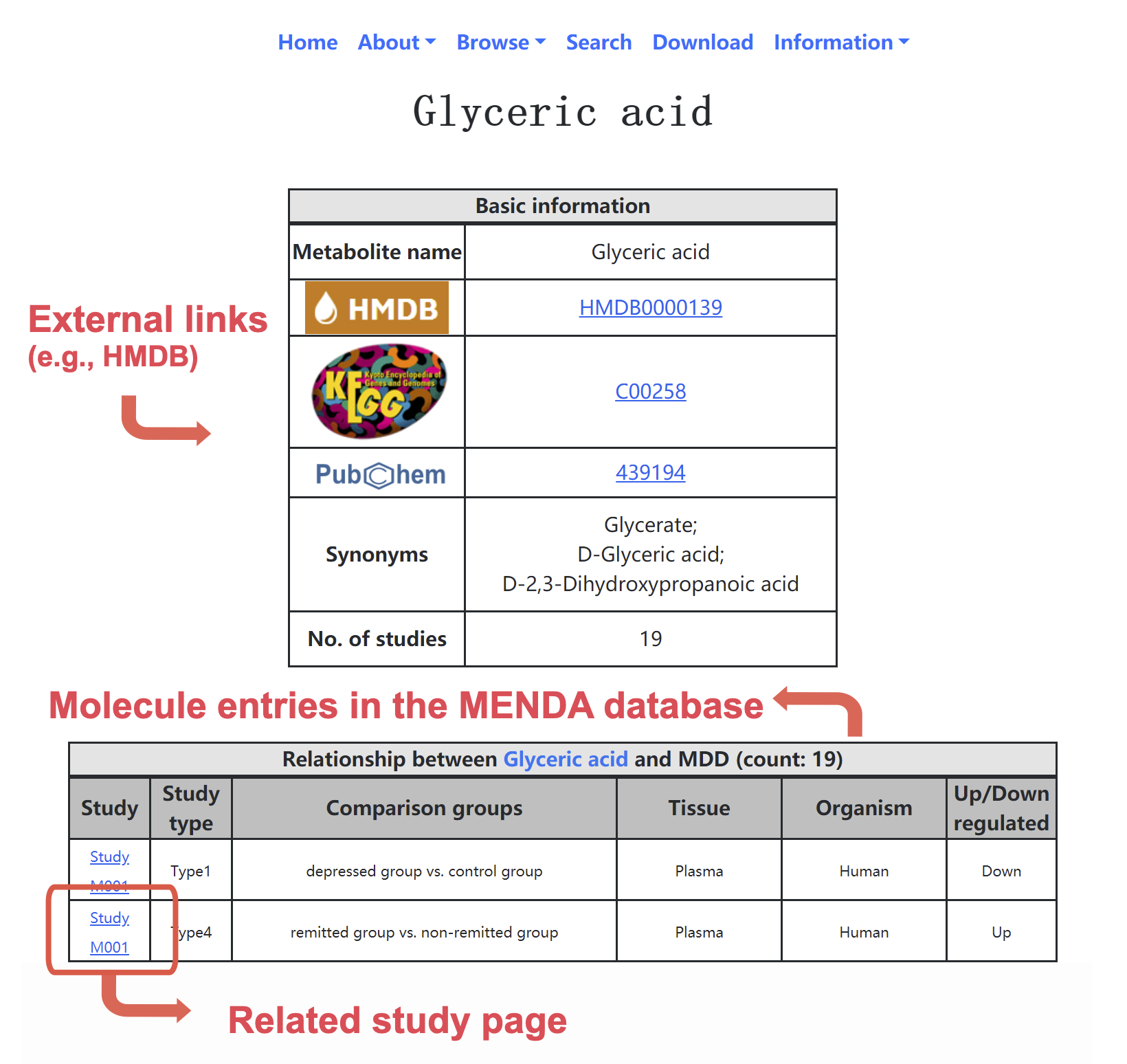
Figure 2. Snapshot of molecule page.
In this database, there are five types of studies that compared metabolite or protein levels between groups.
Type 1 studies are studies that compared molecule levels between depressed and non-depressed groups, which represent the metabolic changes in the pathophysiology of depression. For example, depression patients vs. healthy controls, CUMS model group vs. control group.
Type 2 studies are studies that compared molecule levels between treated depression and non-treated depression groups, which reflect metabolic changes resulted from antidepressant treatments. For example, depression patients with antidepressant group vs. depression patients without antidepressant group, CUMS model + fluoxetine group vs. CUMS model.
Type 3 studies are studies that compared molecule levels between treated and non-treated healthy individuals, which reflect metabolic changes resulted from antidepressant treatments in healthy states. For example, healthy controls with antidepressant group vs. healthy controls without antidepressant group, control rat + fluoxetine group vs. control rat group.
Type 4 studies are studies that compared molecule levels between responder and non-responders of depression after antidepression treatment, which reflect metabolic changes resulted from treatment response in depression states. For example, responders vs. non-responders of fluoxetine-treated patients with depression.
Type 5 studies are studies that compared molecule levels between responder and non-responders of healthy controls after antidepression treatment, which reflect metabolic changes resulted from treatment response in healthy states. For example, responders vs. non-responders of fluoxetine-treated healthy controls.
2) The study-level information
In this part, individual studies are listed numerically. For each study, the information of interest (including study title, number of related molecules, study type, organism, category of depression, tissue, platform) are provided. Search, filter, select and download functions are also provided for the study-level information.
• Study page: Users can click the link of the study IDs, then jump to the specified page for this study. In the specified page (Figure 3), the information of interest (including study name, title, overall design, study type, data availability, organism, category of depression, criteria for depression, sample size, tissue, platform, PMID, and citation) are provided. Molecule entries are also listed.
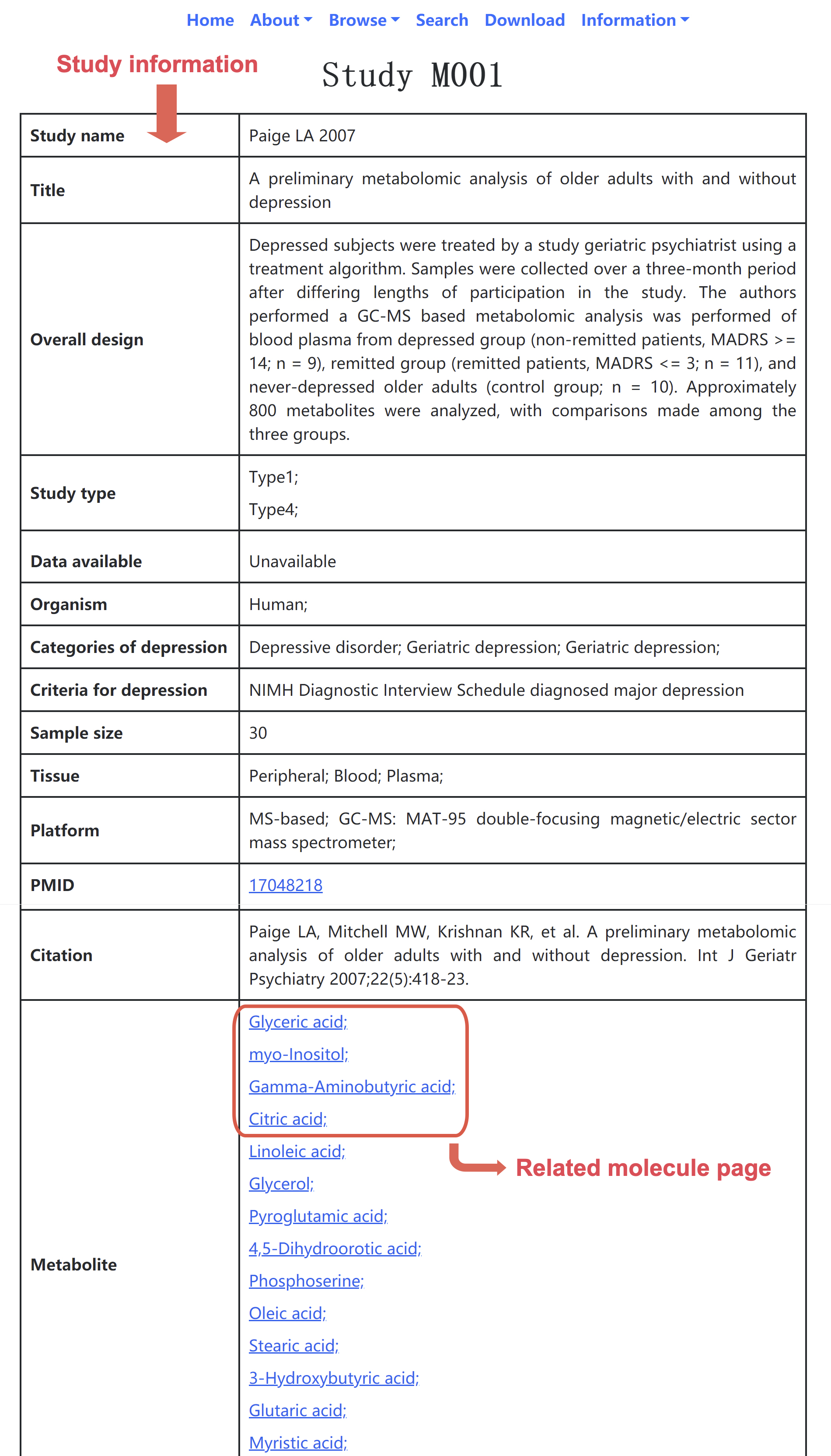
Figure 3. Snapshot of study page.
3. Search
In the search page (Figure 4), users can search individual metabolites using metabolite names or standard identifiers (HMDB, KEGG, or PubChem IDs), or search individual proteins using protein names, gene symbols or standard identifiers (UniProt accessions or UniProt entry names).
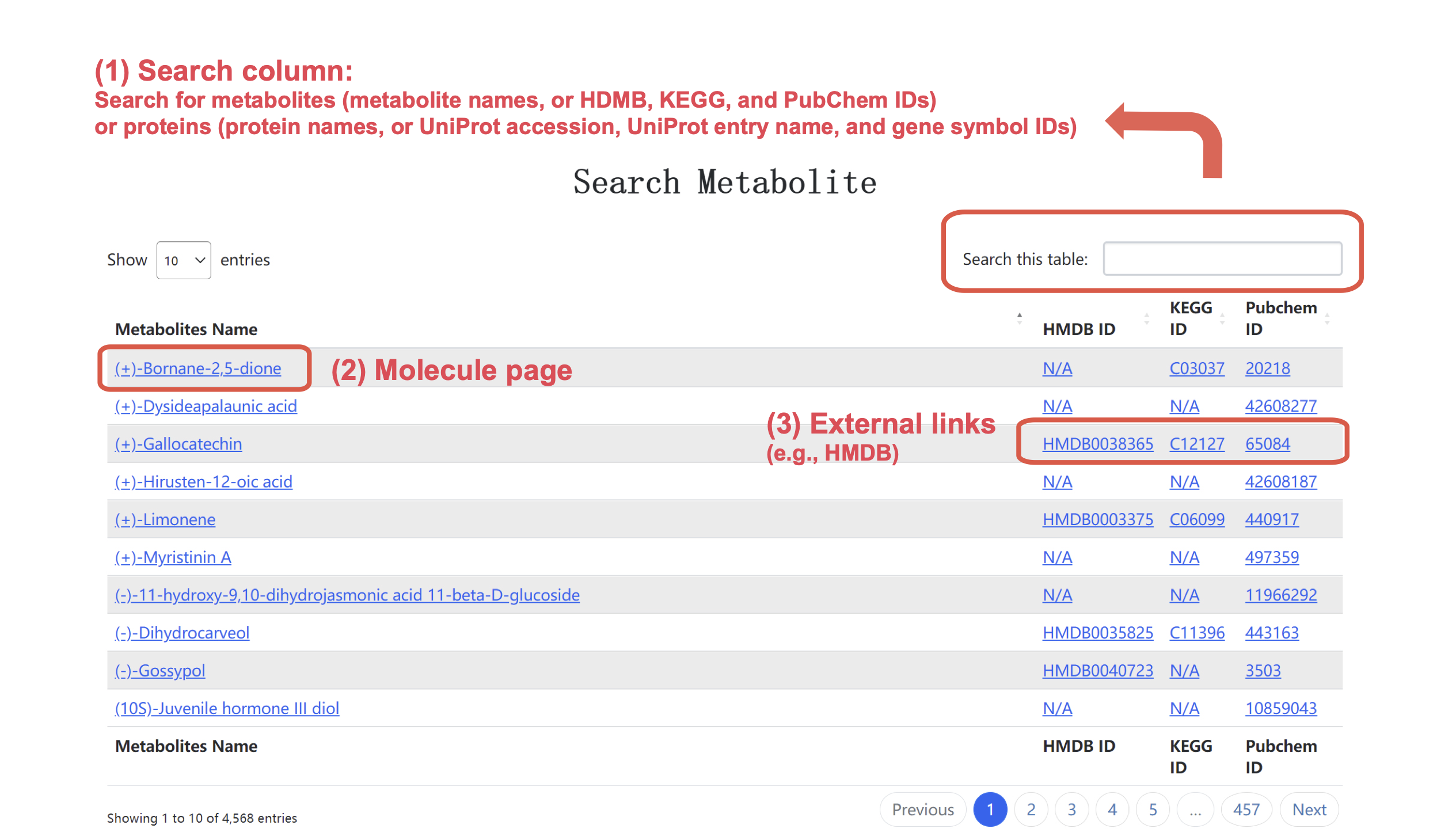
Figure 4. Snapshot of search page.
4. Download
Users can download the core dataset in ProMENDA.