Introduction to ProMENDA
ProMENDA (Protein and Metabolite Network of Depression Database) is a comprehensive database that integrated all available knowledge of metabolomics and proteomics studies for depression. In the following section, we will introduce the details of the ProMENDA database.
2. Data Extraction and Data Annotation
1. Data Collection
1.1 Literature Search
Five electronic databases, including PubMed, Cochrane Library, EMBASE, Web of science, and PsycINFO, were searched.
• PubMed: PubMed is a free search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics provided by the United States National Library of Medicine.
• Cochrane Library: Cochrane Library is a collection of databases in medicine and other healthcare specialties provided by Cochrane and other organizations.
• EMBASE: EMBASE is a biomedical and pharmacological database of published literature designed to support information managers and pharmacovigilance in complying with the regulatory requirements of a licensed drug produced by Elsevier.
• Web of science: Web of science is an online subscription-based scientific citation indexing service originally produced by the Institute for Scientific Information.
• PsycINFO: PsycINFO is a database of abstracts of literature in the field of psychology produced by the American Psychological Association.
Further relevant studies were obtained by manual search of reference lists of all included studies identified in the initial search. The citation lists were also screened in Google Scholar.
1.2 Repository Search
Five metabolomics databases, including Human Metabolome Database (HMDB), MetaboLights, Metabolomics Workbench, MetabolomeXchange, and Omics Discovery Index, were also searched by browsing studies or by imprecise search terms.
• HMDB: HMDB is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in the human body created by the Human Metabolome Project funded by Genome Canada.
• MetaboLights: is a data repository founded in 2012 for cross-species and cross-platform metabolomic studies that provides primary research data and meta data for metabolomic studies as well as a knowledge base for properties of individual metabolites maintained by the European Bioinformatics Institute.
• Metabolomics Workbench: Metabolomics Workbench is a public repository for metabolomics metadata and experimental data spanning various species and experimental platforms, metabolite standards, metabolite structures, protocols, tutorials, and training material and other educational resources sponsored by the United States National Institutes of Health.
• MetabolomeXchange: MetabolomeXchange is an international data aggregation and notification service for metabolomics developed by the Analytical BioSciences group/Leiden University.
• Omics Discovery Index: Omics Discovery Index is a data repository to provide dataset discovery across a heterogeneous, distributed group of Transcriptomics, Genomics, Proteomics and Metabolomics data resources.
1.3 Study Selection Criteria
• Study type: The following five types of studies were included:
Type 1: Depressed group versus control group. The aim of this type of studies was to identify candidate molecules between depressed and healthy states.
Type 2: Treated depressed group versus untreated depressed group. The aim of this type of studies was to identify candidate molecules resulting from treatments in the depressed state.
Type 3: Treated healthy group versus untreated healthy group. The aim of this type of studies was to identify candidate molecules resulting from treatments in the healthy state.
Type 4: Responder versus non-responder in treated depressed group. The aim of this type of studies was to identify candidate molecules resulting from treatment response in the depressed state.
Type 5: Responder versus non-responder in treated healthy group. The aim of this type of studies was to identify candidate molecules resulting from treatment response in the healthy state.
• Organism: Both depressed patients (with depressive disorders or depressive symptoms) and animal models were included. Transgenic animals were excluded.
• Tissue: All types of tissues in the central system (e.g., prefrontal cortex, hippocampus) or peripheral system (e.g., plasma, liver) are included.
• Platform: For metabolites, nuclear magnetic resonance- and mass spectrometry-based metabolomics technologies, including nuclear magnetic resonance, gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS) and others, were included. In addition, we also included studies that use magnetic resonance spectroscopy, since magnetic resonance spectroscopy is the most commonly used method to quantify metabolic changes in the living brain.
For proteins, all mass spectrometry-based proteomics technologies, including label-based (such as iTRAQ, TMT, etc) and label-free technologies.
• Exclusion criteria: Duplicate studies; other detection methods than those previously described; no control of interest; or other type of reports (review, case report, commentary, study protocol). Where studies included ‘mixed disorders’, we excluded studies involving less than 80% patients with a primary diagnosis of depression. Studies without positive results were not excluded.
2. Data Extraction and Data Annotation
All data were manually extracted using standardized data abstraction spreadsheets by independently researchers.
For study-level data, the following items were extracted and annotated:
•Study ID: The ID of each study (e.g., Study M015, Study P012). ‘M’ stands for metabolomics, and ‘P’ stands for proteomics.
•Study name: The name of each study (the first author’s name plus publication year, e.g., Paige LA 2007).
•Title: The title of each study.
•Overall design: The general information of each study, including experimental objectives and experimental methods.
•Criteria for depression: The criteria for depression patients (e.g., DSM-IV diagnosed MDD, HAMD-17 > 22) or animal models (e.g., sucrose preference test, forced swimming test).
•Sample size: Number of samples used for metabolomics and proteomics analysis.
•Original data availability: If the original data was publicly available, the website link was shown; otherwise, we marked with ‘Unavailable’.
•PMID: The PMID of each study from PubMed.
•DOI: The DOI of each study (please visit http://dx.doi.org/ for resolving a DOI Name).
•Citation: The citation of each study.
For metabolite data, the following items were extracted and annotated:
•Metabolite name: The official name of each metabolite (names from HMDB were preferred).
•HMDB: The HMDB ID of each metabolite.
•KEGG: The KEGG ID of each metabolite.
•Pubchem: The Pubchem ID of each metabolite.
•Synonyms: The common synonyms of each metabolite.
•Groups: The comparison groups for each metabolite (e.g., depression group vs. control group).
•Up/Down regulated: The metabolite was up- or down-regulated.
For protein data, the following items were extracted and annotated:
•Protein name: The official name of each protein (names from UniProt were preferred).
•UniProt accession: The UniProt ID of each protein.
•UniProt entry name: The UniProt name of each metabolite.
•Gene symbol: Official gene symbol of each protein.
•Synonyms: The common synonyms of each protein.
•Groups: The comparison groups for each protein (e.g., depression group vs. control group).
•Up/Down regulated: The protein was up- or down-regulated.
For both molecule-level and study-level data:
•Study type: Studies were categorized as five types:
Type 1: Depressed group versus control group. The aim of this type of studies was to identify candidate molecules between depressed and healthy states.
Type 2: Treated depressed group versus untreated depressed group. The aim of this type of studies was to identify candidate molecules resulting from treatments in the depressed state.
Type 3: Treated healthy group versus untreated healthy group. The aim of this type of studies was to identify candidate molecules resulting from treatments in the healthy state.
Type 4: Responder versus non-responder in treated depressed group. The aim of this type of studies was to identify candidate molecules resulting from treatment response in the depressed state.
Type 5: Responder versus non-responder in treated healthy group. The aim of this type of studies was to identify candidate molecules resulting from treatment response in the healthy state.
•Organism_1: The original reported organism (e.g., Human, Sprague-Dawley rat).
•Organism_2: Four categories of organism, including Human, Rat, Mouse, and Non-human primate.
•Categories_of_depression_1: The original reported categories (e.g., Chronic systolic heart failure with depression, Chronic mild stress model).
•Categories_of_depression_2: A total of 18 categories so far, including 12 categories of depression or depressive symptom (Depression, Geriatric depression, Pediatric depression, Postmenopausal depression, Postpartum depression, Post-stroke depression, Antenatal depressive disorder, Treatment-resistant depression, Depression with comorbidity, Depression with suicide, Depressive symptom, and Depressive symptom with comorbidity), and 6 categories of animal models (including the five most commonly used animal models, namely Chronic mild stress model, Social defeat stress model, Chronic restraint stress model, Learned helplessness model, Lipopolysaccharide induced depression model, and Other animal models), and Healthy individuals.
•Categories_of_depression_3: Four categories, including Depression, Depressive symptom, Animal models, and Healthy individuals.
•Tissue_1: The original reported tissue (e.g., Hippocampus, Plasma).
•Tissue_2: A total of 20 categories so far, including Brain, Cerebrospinal fluid, Spinal cord, Adipose tissue, Adrenal gland, Blood, Faece, Fibroblast, Gut, Hair, Heart, Kidney, Liver, Muscle, Ovary, Saliva, Tear, Testis, Thyroid, and Urine.
•Tissue_3: Two categories of tissue, including Central system and Peripheral system.
•Platform: The omics platform original reported (e.g., NMR: Varian INOVA 600 spectrometer).
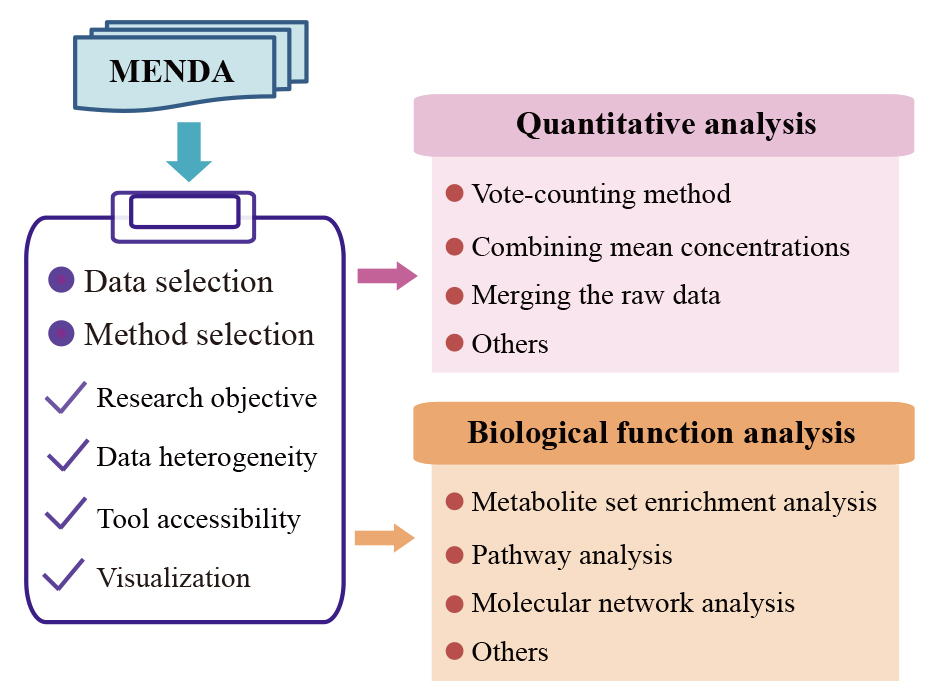
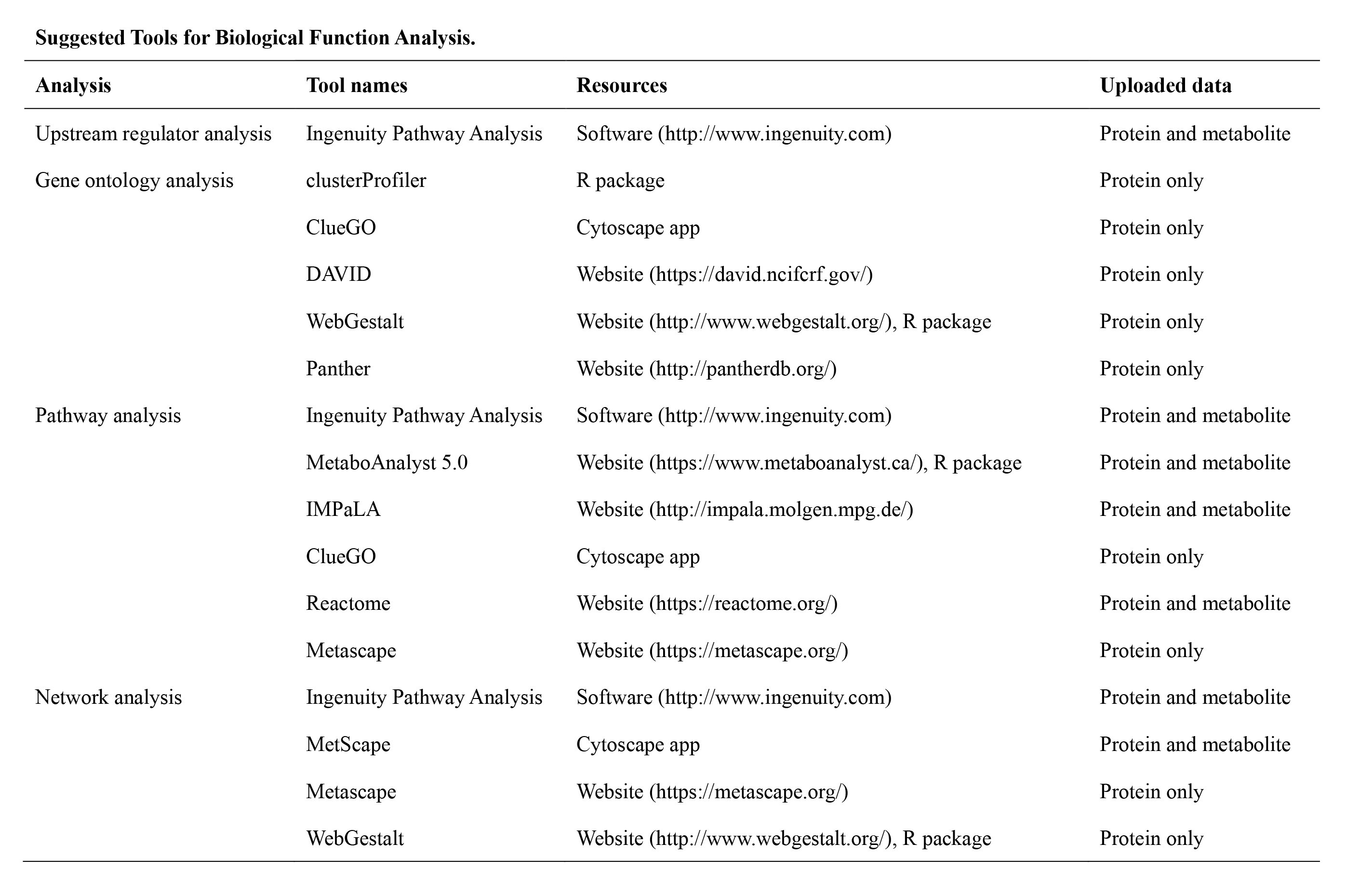
3. Data Analyses
To facilitate data analysis and interpretation for users, we proposed a systematic framework. Users can download the dataset provided in the ProMENDA and perform personalized analysis.